Scientists Develop a New Workflow for Manipulating Protein Chemistry and Structure
New computational tool interprets the chemical changes in proteins, bringing us closer to understanding how to engineer cells.
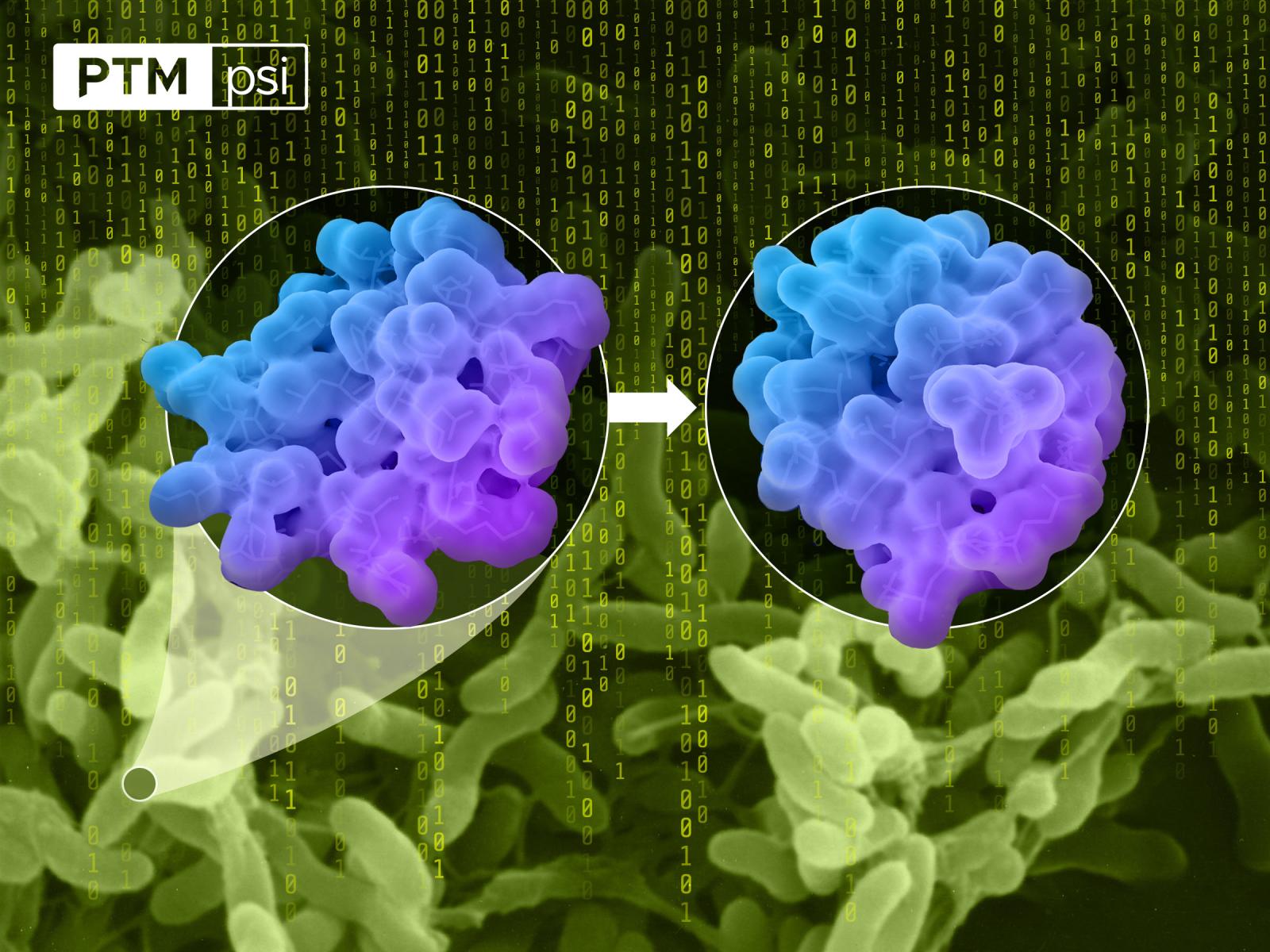
The PTM-Psi generates complex workflows to manipulate protein structures and to interrogate the impact of PTMs on proteins by producing necessary input files and launching instances from more specialized software packages.
(Image by Cortland Johnson | Pacific Northwest National Laboratory)
The Science
Thousands of different proteins play important roles in the human body. Built by strings of amino acids, proteins fold into different shapes and carry out various functions such as cell growth. Proteins are complex because they can change chemically to adapt to their surroundings (known as post-translational modifications or PTMs) and they do so without revealing their exact structure or function. It’s a conundrum that scientists have studied for years because to unravel the structure and function of proteins is to understand how to manipulate cells within humans, animals, and plants. Advancements in computing are bringing us closer to unraveling the mysteries of proteins, but interpreting the vast amounts of data from high-throughput experiments remains a much larger task.
The Impact
We have entered an era where every protein structure can be inferred from its sequence using tools that are driven by artificial intelligence. However, scientists are still far from understanding how these proteins communicate with each other as they change chemically.
A new computational tool developed at Pacific Northwest National Laboratory (PNNL), called PTM-Psi, enables the molecular interpretation of these chemical modifications as a living microbe adapts to its surroundings. Scientists at PNNL aim to connect the critical modifications on the protein structures and the living traits of a microbe, learning the governing principles of protein functions. By reaching the capability to characterize them at a proteomic level, scientists can design protein functions to counter future pandemics, assist bioremediation efforts for plastic waste management, or develop bio-inspired energy technologies.
Summary
The new PTM-Psi workflow opens the door for uncovering molecular insights within proteomics datasets. It does so by integrating several established open-source software packages that consolidate the pertinent information and present it to the user in such a way that they can tease out the pieces of interest. For example, a user will be able to infer a protein’s structure based on its sequence data or calculate free energy perturbations through molecular dynamics simulations.
A workflow like this increases efficiency in data analysis and reduces the time and resources needed to learn or use multiple software packages.
Funding
This work was supported by the Predictive Phenomics Initiative Laboratory Directed Research and Development Program. A portion of the research was performed using the Molecular Sciences Computing Facility at the Environmental Molecular Sciences Laboratory and using resources available through Research Computing at PNNL, a multiprogram national laboratory operated by Battelle for the Department of Energy.
Published: January 2, 2024
Mejia-Rodriguez D, H Kim, N Sadler, X Li, P Bohutskyi, M Valiev, WJ Qian, MS Cheung. 2023. “PTM-Psi: A Python Package to Facilitate the Computational Investigation of Post-Translational Modification on Protein Structures and Their Impacts on Dynamics and Functions.” Protein Sci. 2023; e4822. doi:10.1002/pro.4822