Explicit Representation of Complex Organic Matter Chemistry in Biogeochemical Modeling
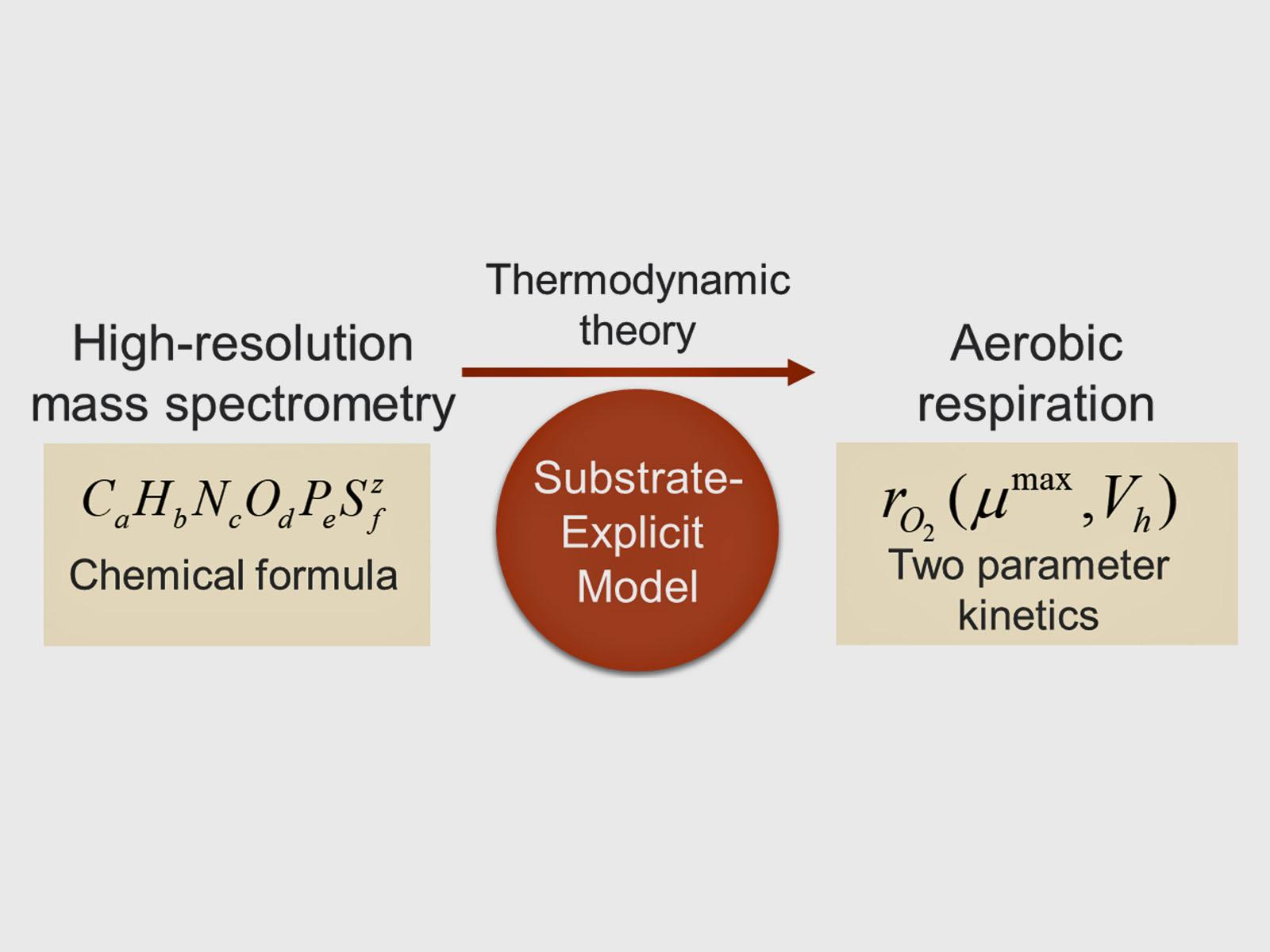
A new framework, called substrate-explicit modeling, uses thermodynamic theory to formulate reaction rate kinetics of organic matter decomposition from chemical formulas.
(Image courtesy of Hyun-Seob Song | University of Nebraska-Lincoln)
The Science
Microbial degradation of organic matter drives biogeochemical cycling on a regional and global scale. However, no modeling framework can capture the molecular details of complex organic matter chemistry in a way that predicts biogeochemical cycling at the ecosystem level; this is mainly due to the difficulty in suitably parameterizing them. Now a team of researchers have developed a new concept of thermodynamics-based biogeochemical modeling that formulates reaction rates in terms of only two parameters regardless of the complexity of organic matter profiles. This approach, termed substrate-explicit modeling, incorporates high-resolution mass spectrometry data for large-scale biogeochemical and reactive transport simulations. In a comparative analysis of two biogeochemically distinct sites, the substrate-explicit model incorporated thousands of compounds to show that thermodynamic properties are a main driver of aerobic respiration.
The Impact
Representing reaction rates based on a limited number of thermodynamic parameters removes a barrier in accounting for complex organic matter chemistry in large-scale ecosystem simulations. This feature can significantly facilitate unprecedented predictions of biogeochemical and ecosystem dynamics, supporting enhanced integration of data and modeling for research programs including the National Ecological Observation Network and the Worldwide Hydrobiogeochemistry Observation Network for Dynamic River Systems (WHONDRS).
Summary
Existing approaches to model organic matter decomposition dynamics can account for details of microbial and/or enzymatic processes to a degree; however, none can yet describe complex organic matter chemistry revealed by high-throughput omics data. This significantly limits understanding of the dynamic interplay between microbes, enzymes, and substrates in biogeochemical cycling.
To fill this gap, scientists from the U.S. Department of Energy’s Pacific Northwest National Laboratory and their collaborators proposed substrate-explicit modeling. This approach incorporates high-resolution metabolomics data to represent complex chemistry based on the thermodynamic properties of organic matter pools. It combines a suite of previously developed thermodynamic theories to characterize reaction kinetics with only two parameters—maximal growth rate and harvest volume—regardless of the number of chemical compounds in an organic matter pool.
The researchers compared predictions from this model with experimental results from two sites with distinct organic matter thermodynamics. The predictions were consistent with their previously reported findings for how thermodynamic properties of an organic matter pool control aerobic respiration across carbon and/or oxygen-limited conditions. The researchers also combined substrate-, microbe-, and enzyme-explicit models to further improve the predictions. The new modeling concept proposed in this work could provide unprecedented data-model integration to be a foundational platform for predictive biogeochemical and ecosystem simulations across scales.
Contact
Tim Scheibe
Pacific Northwest National Laboratory
tim.scheibe@pnnl.gov
Funding
This research was supported by the U.S. Department of Energy (DOE), Biological and Environmental Research program, as part of the Subsurface Biogeochemical Research program. This contribution originates from the Subsurface Biogeochemical Research Scientific Focus Area at the Pacific Northwest National Laboratory and was supported by the partnership with the IDEAS-Watersheds. A portion of the research was performed at the Environmental Molecular Science Laboratory, a DOE Office of Science User Facility located on PNNL’s campus.
Published: December 4, 2020
H.-S. Song, et al., “Representing Organic Matter Thermodynamics in Biogeochemical Reactions via Substrate-Explicit Modeling.” Frontiers in Microbiology 11, 531756 (2020). [DOI: 10.3389/fmicb.2020.531756]