Aaron Wright
Aaron Wright
Biography
Aaron Wright is a senior scientist leading the Biological Systems Science Group in the Biological Sciences Division at PNNL. He has a joint appointment as a research professor in the Gene and Linda Voiland School of Chemical Engineering and Bioengineering at Washington State University (WSU) and an adjunct faculty appointment with the School of Molecular Biosciences at WSU. Wright earned his PhD in organic chemistry from the University of Texas at Austin with professor Eric Anslyn performing research on host-guest sensors for small molecules, peptides, and proteins relevant to biomedical applications. Realizing the potential of synthetic chemistry to answer biological questions, Wright completed a California Breast Cancer Research Program postdoctoral fellowship with professor Benjamin Cravatt at the Scripps Research Institute. While there, his research focused on the general areas of chemical biology and activity-based protein profiling (ABPP), with an emphasis on cytochrome P450 oxidative metabolism.
About the Biological Systems Science Group
Wright’s highly collaborative and diverse chemical biology research team is focused on gaining an improved functional and mechanistic understanding of biological processes including:
- spatiotemporal, functional, and interaction dynamics of microbes and microbiomes;
- oxidative and conjugation metabolism in mammalian liver and lung, particularly with regard to environmental exposures and the effect of developmental stage;
- and the coupling of host metabolism to the gut microbiome.
To improve understanding of biological processes, the team synthesizes chemical probes that are coupled to high sensitivity omics, imaging, and other analytics, in addition to 'traditional' microbiology, genetics, and molecular biology experiments. Wright’s team is a talented composite of biologists, chemists, and chemical biologists and includes research staff, postdoctoral and post-bachelor research associates, graduate students (from WSU), and undergraduate interns working closely with PNNL.
Research Interest
PNNL’s chemical biology research program is focused on:
- characterizing the functional, spatial, and temporal dynamics of microbes and microbial communities that impact community composition and interactions and effect properties, such as resilience, resistance, and productivity;
- determining the function of drug/xenobiotic metabolizing enzymes in the mammalian liver, lung, and gut microbiome as a function of environmental exposure and developmental stage;
- identifying the interplay between host and gut microbiome metabolic activities; and
- developing novel chemical synthesis and chemoproteomic methodologies.
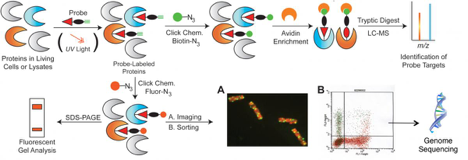
The team develops and deploys active-site, modification-directed, or metabolite-based chemical probes that form irreversible bonds to protein targets in microbes, microbial communities, and mammalian cells and tissues, and subsequently report on probe labeling events by mass spectrometry (MS)-based proteomics, imaging, or flow cytometry. Strengths include: synthetic organic chemistry for probe development, chemoproteomics, environmental, and human health biology. At the simplest level, team members can summarize research as applying chemical probes to living biological systems or cellular extracts, then append enrichment moieties or fluorescent groups via click chemistry for subsequent liquid chromatography mass spectrometry or imaging characterization of probe labeling, (Scheme 1) resulting in an improved functional and mechanistic understanding of biological processes. Below are descriptions of some ongoing projects. Two reviews published in Current Opinion in Chemical Biology and in Biotechnology for Biofuels show the role activity-based protein profiling has on understanding biological functions, interactions, and regulatory dynamics of microbes and microbial communities.
Scheme 1. General strategy for chemical probing of protein functions, interactions, or regulatory modifications. Click chemistry is used to append various reporting moieties to probed proteins for subsequent characterization by imaging, flow cytometry or MS-based proteomics.
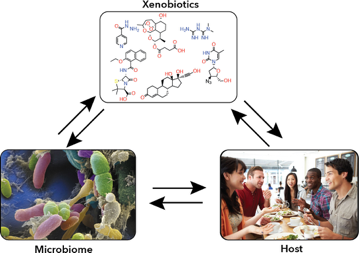
The Gut Microbiome - Drug Metabolism and Exposure Perturbations
The gut microbiome is a key player in human health and development. In particular, gut microbes have an impact on the effect of xenobiotics (foreign chemical compounds) that humans are exposed to throughout life. Many xenobiotics are known to impact the host, but the impact xenobiotics have on the gut microbiome is not well understood. At the same time, gut microbes can also metabolize or alter xenobiotics into different compounds that can have a different impact on the host.
Understanding how host, microbiome, and xenobiotic exposure all interact could guide the development of therapies to mitigate exposure risk or damage. However, the sheer diversity of the gut microbiome makes it difficult to identify the molecular mechanisms underlying specific interactions.
To address this, the team is developing an approach to isolate and identify the microbes and proteins involved in different functions using activity-based probes, and to correlate changes to those functions upon perturbation of the gut microbiome. These small molecules can be used to covalently label only active, functional enzymes within a population.
With the goal to move away from gene- and transcript- based omics and directly measure gut microbiome enzyme activities, the team anticipates providing key information used to develop and guide future therapies. A recent review co-authored with others explores new technologies for characterizing microbiomes (ACS Nano, 2016).
Xenobiotic Metabolism in the Liver and Lung - Ontogeny-Based Function and Response to Xenobiotic Exposure
Scientists from PNNL are engaged in research to understand how oxidative and conjugative (Phase I and II, respectively) metabolic activities in the liver and lung change as a function of age and/or exposures (for example, exposure to polyaromatic hydrocarbons). Specifically, the team is researching organ-level responses to persistent environmental contaminant exposure, drugs, and diet.
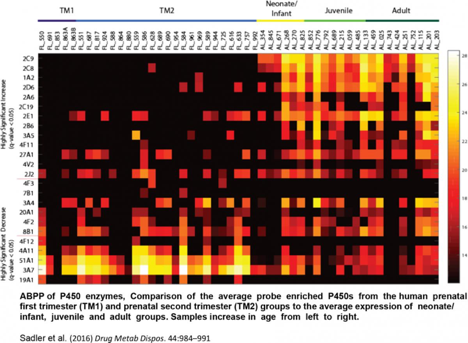
In addition, novel chemical probes for phase II enzymes are being developed and used alongside existing probes for cytochrome P450 enzymes. An initial study determined the effects on P450 functional activity during pregnancy due to exposure to dibenzo[def,p]chrysene in mice (Toxicol Sci. 2013, 135, 48-62). The team followed this study by characterizing human liver P450 activity in samples ranging from prenatal to old age (Drug Metabolism and Disposition, 2016).
Currently, the team is working to submit a manuscript on research exploring perturbations to P450 activity due to the combination of obesity and smoking.
Results will inform pharmacokinetic modeling and will help understand the metabolic and health implications of individual chemicals and environmentally relevant mixtures.
Microbe and Microbiome Functions, Interactions and Spatial Dynamics - Mechanisms for Nutrient Acquisition and Disposition
Nutrient trafficking is a driving force influencing the biogeochemical networks established by microbial communities under metabolic flux. It promotes unique interdependent relationships among microbial members in thriving ecosystems. B-vitamin auxotrophy and opportunism is one of many nutrient-dependent mechanisms with the power to elucidate the evolutionary dynamics behind microbial community structure, resilience, and resistance to perturbations.
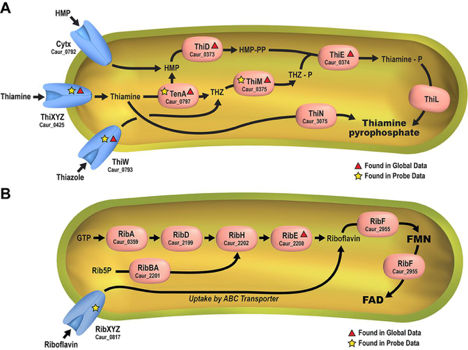
Using a newly synthesized suite of chemical probes derived from B-type vitamins, initial probe efforts led to the experimental validation and identification of numerous substrate-specific B-vitamin transporters and novel intracellular enzyme-cofactor protein associations. Live-cell labeling of the filamentous anoxygenic photoheterotroph, Chloroflexus aurantiacus J-10-fl, was used to employ biosynthetic and salvage-based mechanisms for B-vitamin acquisition. See Figure 2. (ACS Chemical Biology 2016).
Probes provide a unique opportunity to directly link cellular activity and protein function back to native ecosystems and/or host dynamics. The probes do this by targeting protein interactions and vitamin-dependent regulatory functions.
Using tractable probe-based methods in living cells, efforts include innovative experimental designs for directly measuring how microbes respond and adapt to multiple nutrient conditions. These include fluctuations in light intensity, C and N limitation, vitamin deficiency, and carbohydrate utilization in a static biofilm formation. State-of-the-art bioreactor and flow-cell culture techniques are also used.
Live-cell activity and affinity-based probes to allow for multimodal downstream techniques that generate high-throughput functional protein data. These probes are parallel to relative global abundance, transcriptomics, confocal laser microscopy, and cell-sorting techniques. They accurately assign function and specificity for a wide range of experimentally unidentified and predicted membrane-embedded transport proteins, as well as the functional characterization of the intracellular vitamin cofactor-dependent pathways they impact.
Soil Microbiomes - Responding to Environmental Change
Soil microbiomes are essential to environmental nutrient cycling, yet their response to perturbations driven by climate change are still incredibly vague.
The team is investigating the roles hydraulic connectivity and spatial structure have on niche differentiation in the context of substrate availability, the distribution of taxa in soil micro-habitats, and the metabolic capacity of community members. An activity-based protein profiling will be deployed to target enzymes responsible for organic matter decomposition related to nutrient cycling in the environment. This will tease out community member function in varied conditions. These experiments will be used to inform predictive models of soil microbiomes. This work builds off of prior experience in characterizing cellulose and protein degradation—for example, Ser and Cys proteases (J. Am. Chem. Soc. 2012, 134, 20521-32; Molecular BioSystems, 2013, 9, 2992-3000). A project funded by the U.S. Department of Energy, Office of Science, Biological and Environmental Research program will evaluate soil microbial community mechanisms for C acquisition, how these alter community composition and function, and how environmental perturbations impact the communities.
Redox-Based Protein Profiling in Photoautotrophic Cyanobacteria to Characterize Dynamics Associated with Biofuel Production
The team is studying the importance of redox regulation and dynamics on cysteine thiols of the proteins involved in key photosynthetic and metabolic modules within the photoautotrophic cyanobacteria, Synechococcus sp. PCC 7002 and Cyanothece sp. 51142.
Cysteine thiols are susceptible to a range of oxidative modifications, which make them a powerful chemical agent for signaling and regulatory processes in microbes. Additionally, cysteine thiol reduction and oxidation (redox) is a common mechanism used by microbes to sense environmental perturbations and initiate responses.
Photosynthetic cyanobacteria are of great interest because of their potential to generate biofuels. But the regulatory redox dynamics of protein cysteine thiols must first be understood in order to fully realize their capacity.
Proteins involved in critical metabolic pathways, including those important to the synthesis of high-value small molecules, biofuel precursors, and/or hydrogen are hypothesized to have widespread regulation. A critical caveat to investigating thiol redox status is the limitation imposed by cell lysis, in which near-immediate oxidation upon lysis destroys the native redox status.
The team has developed novel chemical probe strategies for targeting reduced protein cysteine thiols in living cells (Frontiers in Microbiology 2014,5:325; ACS Chemical Biology 2014, 9, 291-300; Molecular Carcinogenesis 2014 ,Epub ahead of print, PMID:24285572). An existing chemical probe was used to target sulfenic acid formation.
Thus far, the team has investigated redox dynamics in multiple nutrient conditions, including C and N limitation, high light, and oxidative stress. They have also been able to identify changes to cysteine thiol redox status in as little as 30 seconds in living cells following an environmental perturbation.
By parsing proteins that undergo redox reactions and those highly sensitive to reactive oxygen species stress from the rest of the proteome, a fuller understanding of photoautotrophic regulatory dynamics can be achieved. To validate and extend our redox probing, we have also carried out complementary global proteomic, transcriptomic, confocal laser microscopy (See figure 3), and photophysiology measurements.
In total, this comprehensive approach provides a more thorough understanding of redox signaling that will facilitate optimization of photosynthetic cellular output. Work in this area was covered in Chemical & Engineering News.
Activity-Based Protein Profiling for Lignocellulose-Based Biofuel Applications
Microbial degradation and fermentation of lignocellulosic biomass is a key strategy being investigated in renewable energy research. Cellulolytic microbes and microbial communities are capable of producing β-glucosidases, endo- and exoglucanases, and other glycoside hydrolases to facilitate the degradation of highly recalcitrant cellulose and related plant cell wall polysaccharides. Towards this end, the team developed a large suite of activity-based probes for identifying the functional enzymes involved in cellulose deconstruction published in the J. Am. Chem. Soc, 2012. This suite of probes has been applied to characterize the functional assemblage of the cell-adherent cellulosome structure in the anaerobic bacterium Clostridium thermocellum, and to rapidly identify alterations to cellulolytic activity upon industry- and biorefinery-relevant perturbations in the established biofuel platform production fungi Trichoderma reesei (Molecular Biosystems, 2013).
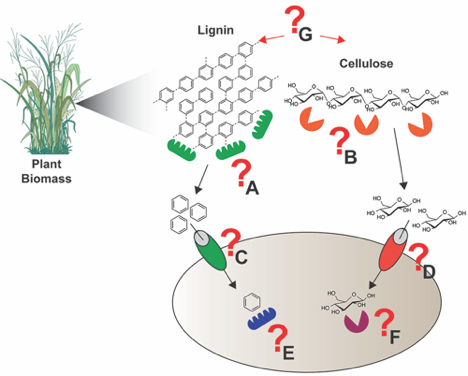
Most recently, they have worked closely with Michelle O’Malley of University of California, Santa Barbara to perform proteomic analyses of fungal lignocellulose degraders. This research resulted in a publication in Science 2016 characterizing a fungal cellulosome system. A follow-on study is in review. Below, Figure 3, shows a general schematic of how we want to use chemical probes and other experiments to understand microbe and microbiome lignocellulose degrading activities. This work ties in with the soil studies described above.
Figure 3. To elucidate and annotate the concert of lignocellulose deconstruction, catabolite transport, and intracellular metabolic activities in microbes and microbiomes we develop ABPP approaches coupled to other experimental techniques. To understand lignocellulose deconstruction we can measure extracellular oxidative lignin depolymerizing enzymes (A) and cellulose degrading enzymes (B), the transport mechanisms for aromatic (C) and carbohydrate (D) catabolites, and the intracellular metabolic activities associated with aromatic (E) and carbohydrate (F) catabolites. Critical associations (G) between lignin depolymerization and cellulose degradation are also of high interest to us, as our microbe-microbe interactions that facilitate lignocellulose deconstruction (Pill/Pac-Man shapes indicate proteins and enzymes).
Education
- PhD in Organic Chemistry, University of Texas, 2006
- BS in Chemistry, George Fox University, 2001
- National Science Foundation’s Research Experience for Undergraduates, University of Washington, 2000
Affiliations and Professional Service
- American Chemical Society
- Society of Industrial Microbiology and Biotechnology
Awards and Recognitions
- Young Investigator Award, American Chemical Society
- Postdoctoral Fellow, California Breast Cancer Research Program
- Graduate Research Fellow, Dorothy Banks Scholarship
- Career Development Grant, University of Texas
- Dick van Santen Award, American Chemical Society
- Edwards-Holman Science Award
- Research Experience for Undergraduates Fellow, The University of Washington, National Science Foundation
Publications
2021
- Wright A.T. 2021. "A transcriptional relationship with a natural product disrupts mitochondrial biogenesis." Cell Chemical Biology 28, no. 10:1392 - 1393. PNNL-SA-166818. doi:10.1016/j.chembiol.2021.10.002
- Stoddard E.G., S. Nag, J. Martin, K.J. Tyrrell, T.M. Gibbins, K.A. Anderson, and A.K. Shukla, et al. 2021. "Exposure to an environmental mixture of polycyclic aromatic hydrocarbons induces hepatic cytochrome P450 enzymes in mice." Chemical Research in Toxicology 34, no. 9:2145 - 2156. PNNL-SA-163871. doi:10.1021/acs.chemrestox.1c00235
- Jarsberg L.G., K. Kedia, T.G. Wendler, A.T. Wright, P.D. Piehowski, M.A. Gritsenko, and T. Shi, et al. 2021. "Nutritional markers and proteome in patients undergoing treatment for pulmonary tuberculosis differ by geographic region." PLOS ONE 16, no. 5:Article No. e0250586. PNNL-SA-156554. doi:10.1371/journal.pone.0250586
- Swift C., K.B. Louie, B.P. Bowen, H.M. Olson, S.O. Purvine, A. Salamov, and S.J. Mondo, et al. 2021. "Anaerobic gut fungi are an untapped reservoir of natural products." Proceedings of the National Academy of Sciences of the United States of America 118, no. 18:e2019855118. PNNL-SA-160632. doi:10.1073/pnas.2019855118
- Zegeye E., N.C. Sadler, G.X. Lomas, I.K. Attah, J.K. Jansson, K.S. Hofmockel, and C.R. Anderton, et al. 2021. "Activity-based protein profiling of chitin catabolism." Chembiochem 22, no. 4:717-723. PNNL-SA-156004. doi:10.1002/cbic.202000616
- Schultz K.J., S.M. Colby, V.S. Lin, A.T. Wright, and R.S. Renslow. 2021. "Ligand- and Structure-Based Analysis of Deep Learning-Generated Potential alpha2a Adrenoceptor Agonists." Journal of Chemical Information and Modeling 61, no. 1:481-492. PNNL-SA-155859. doi:10.1021/acs.jcim.0c01019
2020
- Sveistyte A., T.M. Gibbins, K.J. Tyrrell, C.J. Miller, M.H. Foley, A.E. Plymale, and A.T. Wright, et al. 2020. "Simple analysis of primary and secondary bile salt hydrolysis in mouse and human gut microbiome samples using fluorogenic substrates." Chembiochem 21, no. 24:3539-3543. PNNL-SA-152472. doi:10.1002/cbic.202000370
- Killinger B.J., V.A. Petyuk, and A.T. Wright. 2020. "Detecting differential protein abundance by combining peptide level P-values." Molecular Omics 16, no. 6:554-562. PNNL-SA-144005. doi:10.1039/d0mo00045k
- Lin V.S., R.F. Volk, A.J. DeLeon, L.N. Anderson, S.O. Purvine, A.K. Shukla, and H. Bernstein, et al. 2020. "Structure dependent determination of organophosphate targets in mammalian tissues using activity-based protein profiling." Chemical Research in Toxicology 33, no. 234:414-425. PNNL-SA-146869. doi:10.1021/acs.chemrestox.9b00344
- Steiger A.K., S.J. Fansler, C. Whidbey, C.J. Miller, and A.T. Wright. 2020. "Probe-Enabled Approaches for Function-Dependent Cell Sorting and Characterization of Microbiome Subpopulations." In Chemical Tools for Imaging, Manipulating, and Tracking Biological Systems: Diverse Methods for Prokaryotic and Eukaryotic Systems: Methods in Enzymology, edited by D.M. Chenoweth. 89-107. San Diego, California:Academic Press. PNNL-SA-150390. doi:10.1016/bs.mie.2020.03.014
2019
- Brandvold K.R., J.M. Weaver, C. Whidbey, and A.T. Wright. 2019. "A continuous fluorescence assay for simple quantification of bile salt hydrolase activity in the gut microbiome." Scientific Reports 9. PNNL-SA-133128. doi:10.1038/s41598-018-37656-7
- Dickinson M., L.N. Anderson, B.M. Webb-Robertson, J.R. Hansen, R.D. Smith, A.T. Wright, and K. Hybiske. 2019. "Proximity-dependent proteomics of the Chlamydia trachomatis inclusion membrane reveals functional interactions with endoplasmic reticulum exit sites." PLoS Pathogens 15, no. 4:Article Number e1007698. PNNL-SA-132851. doi:10.1371/journal.ppat.1007698
- Whidbey C., N.C. Sadler, R.N. Nair, R.F. Volk, A.J. DeLeon, L.M. Bramer, and S.J. Fansler, et al. 2019. "A Probe-Enabled Approach for the Selective Isolation and Characterization of Functionally Active Subpopulations in the Gut Microbiome." Journal of the American Chemical Society 141, no. 1:42-47. PNNL-SA-127532. doi:10.1021/jacs.8b09668
- Wright A.T. 2019. "Gut commensals make choline too." Nature Microbiology 4, no. 1:4-5. PNNL-SA-139558. doi:10.1038/s41564-018-0325-1
2018
- Kedia K., J.P. Wendler, E.M. Baker, K.E. Burnum-Johnson, L.G. Jarsberg, K.G. Stratton, and A.T. Wright, et al. 2018. "Application of multiplexed ion mobility spectrometry towards the identification of host protein signatures of treatment effect in pulmonary tuberculosis." Tuberculosis 112. PNNL-SA-137966. doi:10.1016/j.tube.2018.07.005
- Ortega C., A. Frando, B.M. Webb-Robertson, L.N. Anderson, N. Fleck, E.L. Flannery, and M. Fishbaugher, et al. 2018. "A global survey of ATPase activity in Plasmodium falciparum asexual blood stages and gametocytes." Molecular and Cellular Proteomics 17, no. 1:111-120. PNNL-SA-121236. doi:10.1074/mcp.RA117.000088
- Sadler N.C., B.M. Webb-Robertson, T. Clauss, J.G. Pounds, R.A. Corley, and A.T. Wright. 2018. "High-fat diets alter the modulatory effects of xenobiotics on cytochrome P450 activities." Chemical Research in Toxicology 31, no. 5:308-318. PNNL-SA-126308. doi:10.1021/acs.chemrestox.8b00008
- Stoddard E.G., R.F. Volk, J.P. Carson, C.M. Ljungberg, T.A. Murphree, J.N. Smith, and N.C. Sadler, et al. 2018. "Multifunctional Activity-Based Protein Profiling of the Developing Lung." Journal of Proteome Research 17, no. 8:2623-2634. PNNL-SA-132239. doi:10.1021/acs.jproteome.8b00086
- Whidbey C., and A.T. Wright. 2018. "Activity-based protein profiling - enabling multimodal functional studies of microbial communities." In Activity-Based Protein Profiling: Current Topics in Microbiology and Immunology", edited by B.F. Cravatt, K.L. Hsu, and E. Weerapana. 1-21. Cham:Springer. PNNL-SA-136518. doi:10.1007/82_2018_128
2017
- Haitjema C., S.P. Gilmore, J.K. Henske, K.V. Solomon, R.D. Groot, A. Kuo, and S.J. Mondo, et al. 2017. "A Parts List for Fungal Cellulosomes Revealed by Comparative Genomics." Nature Microbiology 2. PNNL-SA-120227. doi:10.1038/nmicrobiol.2017.87
- Nair R.N., J.J. Rosnow, T.A. Murphree, M.E. Bowden, S.R. Lindemann, and A.T. Wright. 2017. "De novo synthesis of alkyne substituted tryptophans as chemical probes for protein profiling studies." Organic Chemistry Frontiers 4, no. 4:495-499. PNNL-SA-122250. doi:10.1039/C6QO00819D
- Romine M.F., D.A. Rodionov, Y. Maezato, L.N. Anderson, P. Nandhikonda, I.A. Rodionova, and A. Carre, et al. 2017. "Elucidation of new roles for vitamin B12 in regulation of folate, ubiquinone, and methionine metabolism." Proceedings of the National Academy of Sciences of the United States of America 114, no. 7:E1205-E1214. PNNL-SA-119181. doi:10.1073/pnas.1612360114
- Smith J.N., K.J. Tyrrell, J.R. Hansen, D.G. Thomas, T.A. Murphree, A.K. Shukla, and T. Luders, et al. 2017. "Plasma Protein Turnover Rates in Rats Using Stable Isotope Labeling, Global Proteomics, and Activity-Based Protein Profiling." Analytical Chemistry 89, no. 24:13559-13566. PNNL-SA-129561. doi:10.1021/acs.analchem.7b03984
- Stoddard E.G., B.J. Killinger, R.N. Nair, N.C. Sadler, R.F. Volk, S.O. Purvine, and A.K. Shukla, et al. 2017. "Activity-Based Probes for Isoenzyme- and Site-Specific Functional Characterization of Glutathione S- Transferases." Journal of the American Chemical Society 139, no. 45:16032-16035. PNNL-SA-127421. doi:10.1021/jacs.7b07378
2016
- Anderson L.N., P.K. Koech, A.E. Plymale, E.V. Landorf, A. Konopka, F. Collart, and M.S. Lipton, et al. 2016. "Live Cell Discovery of Microbial Vitamin Transport and Enzyme-Cofactor Interactions." ACS Chemical Biology 11, no. 2:345-354. PNNL-SA-108145. doi:10.1021/acschembio.5b00918
- Bennett K., N.C. Sadler, A.T. Wright, C. Yeager, and M.R. Hyman. 2016. "Activity-based protein profiling of ammonia monooxygenase in Nitrosomonas europaea." Applied and Environmental Microbiology 82, no. 8:2270-2279. PNNL-SA-114336. doi:10.1128/AEM.03556-15
- Biteen J.S., P.C. Blainey, Z.G. Cardon, M. Chun, G. Church, P.C. Dorrestein, and S.E. Fraser, et al. 2016. "Tools for the Microbiome: Nano and Beyond." ACS Nano 10, no. 1:6-37. PNNL-SA-115178. doi:10.1021/acsnano.5b07826
- Ortega C., L.N. Anderson, A. Frando, N.C. Sadler, R.W. Brown, R.D. Smith, and A.T. Wright, et al. 2016. "Systematic Survey of Serine Hydrolase Activity in Mycobacterium tuberculosis Defines Changes Associated with Persistence." Cell Chemical Biology 23, no. 2:290-298. PNNL-SA-112430. doi:10.1016/j.chembiol.2016.01.003
- Perez D.M., M.E. Richards, R. Parker, M.E. Berres, A.T. Wright, M. Sifri, and N.C. Sadler, et al. 2016. "Role of cytochrome P450 hydroxylase in the decreased accumulation of vitamin E in muscle from turkeys compared to that from chickens." Journal of Agricultural and Food Chemistry 64, no. 3:671-680. PNNL-SA-108079. doi:10.1021/acs.jafc.5b05433
- Sadler N.C., H.C. Bernstein, M.R. Melnicki, M.A. Charania, E.A. Hill, L.N. Anderson, and M.E. Monroe, et al. 2016. "Dinitrogenase Driven Photobiological Hydrogen Production Combats Oxidative Stress in Cyanothece sp. ATCC 51142." Applied Environmental Microbiology 82, no. 24:7227-7235. PNNL-SA-113160. doi:10.1128/AEM.02098-16
- Sadler N.C., P. Nandhikonda, B.M. Webb-Robertson, C. Ansong, L.N. Anderson, J.N. Smith, and R.A. Corley, et al. 2016. "Hepatic cytochrome P450 activity, abundance, and expression throughout human development." Drug Metabolism and Disposition 44, no. 7:984-91. PNNL-SA-112422. doi:10.1124/dmd.115.068593
- Solomon K.V., C. Haitjema, J.K. Henske, S.P. Gilmore, D. Borges-Rivera, A. Lipzen, and H.M. Brewer, et al. 2016. "Early-branching Gut Fungi Possess A Large, And Comprehensive Array Of Biomass-Degrading Enzymes." Science 351, no. 6278:1192-1195. PNNL-SA-115073. doi:10.1126/science.aad1431
- Wright A.T. 2016. "HOST-PATHOGEN INTERACTIONS: A cholera surveillance system." Nature Chemical Biology 12, no. 4:203-204. PNNL-SA-115724. doi:10.1038/nchembio.2039
2015
- Bernstein H.C., M.A. Charania, R.S. McClure, N.C. Sadler, M.R. Melnicki, E.A. Hill, and L.M. Markillie, et al. 2015. "Multi-omic Dynamics Associate Oxygenic Photosynthesis with Nitrogenase-mediated H2 production in Cyanothece sp. ATCC 51142." Scientific Reports 5. PNNL-SA-111925. doi:10.1038/srep16004
- Liu Y., J.K. Fredrickson, N.C. Sadler, P. Nandhikonda, R.D. Smith, and A.T. Wright. 2015. "Advancing Understanding of Microbial Bioenergy Conversion Processes by Activity-Based Protein Profiling." Biotechnology for Biofuels 8. PNNL-SA-107650. doi:10.1186/s13068-015-0343-7
- Sadler N.C., and A.T. Wright. 2015. "Activity-Based Protein Profiling of Microbes." Current Opinion in Chemical Biology 24. PNNL-SA-105009. doi:10.1016/j.cbpa.2014.10.022
- Wright A.T., T. Magnaldo, R.L. Sontag, L.N. Anderson, N.C. Sadler, P.D. Piehowski, and Y. Gache, et al. 2015. "Deficient Expression of Aldehyde Dehydrogenase 1A1 Is Consistent with Increased Sensitivity of Gorlin Syndrome Patients to Radiation Carcinogenesis." Molecular Carcinogenesis 54, no. 6:473-484. PNNL-SA-95210. doi:10.1002/mc.22115
2014
- Ansong C., N.C. Sadler, E.A. Hill, M.P. Lewis, E.M. Zink, R.D. Smith, and A.S. Beliaev, et al. 2014. "Characterization of protein redox dynamics induced during light-to-dark transitions and nutrient limitation in cyanobacteria." Frontiers in Microbiology 5. PNNL-SA-101966. doi:10.3389/fmicb.2014.00325
- Ismail H.M., P.M. O'Neill, D. Hong, R. Finn, C. Henderson, A.T. Wright, and B. Cravatt, et al. 2014. "Pyrethroid Activity-Based Probes for Profiling Cytochrome P450 Activities Associated with Insecticide Interactions." Proceedings of the National Academy of Sciences of the United States of America 110, no. 49:19766-19771. PNNL-SA-98703. doi:10.1073/pnas.1320185110
- Liu Y., R. Zhang, Z. Lian, S. Wang, and A.T. Wright. 2014. "Yeast cell surface display for lipase whole cell catalyst and its applications." Journal of Molecular Catalysis B: Enzymatic 106. PNNL-SA-102321. doi:10.1016/j.molcatb.2014.04.011
- Ortega C., R. Liao, L.N. Anderson, T. Rustad, A.R. Ollodart, A.T. Wright, and D.R. Sherman, et al. 2014. "Mycobacterium tuberculosis Ser/Thr protein kinase B mediates an oxygen-dependent replication switch." PLoS Biology 12, no. 1:Article No. e1001746. PNNL-SA-94282. doi:10.1371/journal.pbio.1001746
- Pena-Castillo L., R. Mercer, A. Gurinovich, S.J. Callister, A.T. Wright, A. Westbye, and J.T. Beatty, et al. 2014. "Gene co-expression network analysis in Rhodobacter capsulatus and application to comparative expression analysis of Rhodobacter sphaeroides." BMC Genomics 15. PNNL-SA-105663. doi:10.1186/1471-2164-15-730
- Sadler N.C., M.R. Melnicki, M.H. Serres, E.D. Merkley, W.B. Chrisler, E.A. Hill, and M.F. Romine, et al. 2014. "Live Cell Chemical Profiling of Temporal Redox Dynamics in a Photoautotrophic Cyanobacterium." ACS Chemical Biology 9, no. 1:291-300. PNNL-SA-93812. doi:10.1021/cb400769v
- Wiedner S.D., L.N. Anderson, N.C. Sadler, W.B. Chrisler, V.K. Kodali, R.D. Smith, and A.T. Wright. 2014. "Organelle-Specific Activity-Based Protein Profiling in Living Cells." Angewandte Chemie International Edition 53, no. 11:2919-2922. PNNL-SA-98713. doi:10.1002/anie.201309135
2013
- Anderson L.N., D.E. Culley, B.A. Hofstad, L.M. Chauvigne-Hines, E.M. Zink, S.O. Purvine, and R.D. Smith, et al. 2013. "Activity-based protein profiling of secreted cellulolytic enzyme activity dynamics in Trichoderma reesei QM6a, NG14, and RUT-C30." Molecular Biosystems 9, no. 12:2992-3000. PNNL-SA-97265. doi:10.1039/c3mb70333a
- Ansong C., C. Ortega, S.H. Payne, D.H. Haft, L.M. Chauvigne-Hines, M.P. Lewis, and A.R. Ollodart, et al. 2013. "Identification of widespread adenosine nucleotide binding in Mycobacterium tuberculosis." Chemistry & Biology 20, no. 1:123-133. PNNL-SA-88963. doi:10.1016/j.chembiol.2012.11.008
- Crowell S.R., A. Sharma, S. Amin, J.J. Soelberg, N.C. Sadler, A.T. Wright, and W.M. Baird, et al. 2013. "Impact of Pregnancy on the Pharmacokinetics of Dibenzo[def,p]chrysene in Mice." Toxicological Sciences 135, no. 1:48-62. PNWD-SA-10207. doi:10.1093/toxsci/kft124
- Wiedner S.D., C. Ansong, B.M. Webb-Robertson, L.M. Pederson, S. Fortuin, B.A. Hofstad, and A.K. Shukla, et al. 2013. "Disparate Proteome Responses of Pathogenic and Non-pathogenic Aspergilli to Human Serum Measured by Activity-Based Protein Profiling (ABPP)." Molecular & Cellular Proteomics. MCP 12, no. 7:1791-1805. PNNL-SA-91969. doi:10.1074/mcp.M112.026534
2012
- Chauvigne-Hines L.M., L.N. Anderson, H.M. Weaver, J.N. Brown, P.K. Koech, C.D. Nicora, and B.A. Hofstad, et al. 2012. "Suite of Activity-Based Probes for Cellulose-Degrading Enzymes." Journal of the American Chemical Society 134, no. 50:20521-20532. PNNL-SA-91066. doi:10.1021/ja309790w
- Sadler N.C., T.E. Angel, M.P. Lewis, L.M. Pederson, L.M. Chauvigne-Hines, S.D. Wiedner, and E.M. Zink, et al. 2012. "Activity-Based Protein Profiling Reveals Mitochondrial Oxidative Enzyme Impairment and Restoration in Diet-Induced Obese Mice." PLoS One 7, no. 10:Article No. e47996. PNNL-SA-88011. doi:10.1371/journal.pone.0047996
- Wiedner S.D., K.E. Burnum, L.M. Pederson, L.N. Anderson, S. Fortuin, L.M. Chauvigne-Hines, and A.K. Shukla, et al. 2012. "Multiplexed Activity-based Protein Profiling of the Human Pathogen Aspergillus fumigatus Reveals Large Functional Changes upon Exposure to Human Serum." Journal of Biological Chemistry 287, no. 40:33447-33459. PNNL-SA-87258. doi:10.1074/jbc.M112.394106